

|
| This project is collaborated between The Forsyth Institute (TFI) and The Institute for Genomic Research (TIGR), and is funded by National Institute of Dental and Craniofacial Research (NIDCR) |
|
|
|||||||||||||||||||||||||
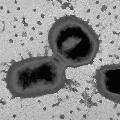
Specific Aims of the Porphyromonas gingivalis Genome ProjectPorphyromonas gingivalis is the organism most strongly associated with human periodontal disease. It is a member of the Cytophaga/Flavobacter/Bacteroides (CFB) phylum of bacteria, a group phylogenetically more distant from Eschericheria coli, Haemophilus influenzae, Pseudomonas aeruginosa, and Helicobacter pylori, than Gram-positive bacteria. Porphyromonas and Bacteroides spp. are very common causes of human infections, and are among a short list of pathogens that are currently causing the greatest antibiotic resistance problems. While the genome size of most members of the CFB phylum are in the 4-6 megabase range, P. gingivalis has a genome of only 2.34 megabases. We propose to sequence the complete genome of Porphyromonas gingivalis because: it is important in periodontal disease; it is a small genome representative of a medically and phylogenetically important but under-studied CFB group of bacteria; and there is a large group of researchers with appropriate genetic tools to utilize the sequence information for elucidating virulence mechanisms and for vaccine development. There are four specific goals for this genome project: |
|||
|
This page is created and maintained by Drs. Margaret Duncan, Floyd Dewhirst, and Tsute Chen, Department of Molecular Genetics, The Forsyth Institute . Last modified on 02/20/2002 Copyright 2000, 2001, 2002 by The Forsyth Institute |