

|
| This project is collaborated between The Forsyth Institute (TFI) and The Institute for Genomic Research (TIGR), and is funded by National Institute of Dental and Craniofacial Research (NIDCR) |
|
|
|||||||||||||||||||||||||
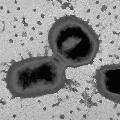
Pathogenesis studiesPorphyromonas gingivalis possesses an armamentarium of cell-surface associated and extracellular activities, which are studied intensively for their virulence potential. Several are putative adhesins which interact with other bacteria, epithelial cells, and extracellular matrix proteins. Secreted or cell-bound enzymes, toxins, and hemolysins may play a significant role in the spread of the organism through tissue, in tissue destruction, and in evasion of host defenses. Three groups reported oral epithelial cell invasion by laboratory and clinical isolates of P. gingivalis (Lamont et al. 1992; Duncan et al.1993; Sandros et al.1993). Recently it was reported that P. gingivalis invasion was accompanied by intracellular calcium-fluxes (Chan et al., 1995) and inhibited by cytochalasin D and nocodazole, indicating that rearrangements in the cytoskeleton of the epithelial cell are necessary for internalization (Lamont et al., 1995). The authors also report that protein kinase signaling pathways within epithelial cells are associated with P. gingivalis invasion as in other systems (Rosenshine et al., 1994). The genes associated with invasion are being investigated. Biochemical, immunological and genetic evidence indicates that P. gingivalis fimbriae are involved in adhesion to both saliva-coated hydroxy-apatite and to human oral epithelial cells (Lee et al., 1992, 1993; Isogai et al., 1988; Watanabe et al., 1992; Duncan et al., 1993; Hamada et al., 1994). Animal studies suggest fimbriae play a role in host colonization (Malek et al., 1994). The fimA gene encoding fimbrillin, the subunit protein of fimbriae, has been cloned from several P. gingivalis strains. Sequence comparison reveals both conserved and variable regions among strains (Dickinson et al., 1988; Fujiwara et al., 1993). Porphyromonas gingivalis possesses three major proteolytic activities with a) trypsin-like, b) collagenolytic, and c) glycylprolyl peptidase activities. Numerous proteins with thiol-dependent trypsin-like cleavage specificity can be isolated from cells and culture supernatants, but biochemical and genetic analyses indicate that many of these are derived by proteolytic processing of a larger, cell-associated, primary gene product (Ciborowski et al., 1994, Pavloff et al., 1995). Structurally, the proteases contain a propeptide sequence, an N-terminal active site and a C-terminal putative adhesin domain with repeat regions. Recent studies indicate that P. gingivalis possesses a family of genes which contains part of the protease sequences (Fletcher et al., 1995; Aduse-Opuku et al., 1995, Nakayama et al., 1995; Potempa et al., 1995; Barkocy-gallagher et al., 1996). These proteases also possess hemagglutinating activity and share extensive DNA sequence homology with hemagglutinin genes hagA and D (A. Progulske-Fox, personal communication; Han et al., 1993; Barkocy-gallagher et al., 1996). The gene for another cysteine protease with different putative adhesive domains has also been cloned (Madden et al., 1994). It has been demonstrated in vitro that P. gingivalis thiol proteases bind to and cleave extracellular matrix proteins, promote vascular permeability, disrupt polymorphonuclear leukocyte function, cleave complement, and degrade IgG heavy chains (Lantz et al., 1991; Imamura et al., 1994; Nakayama et al., 1995; Schenkein, 1988; Killian, 1981). Considerable effort by many groups is being expended isolating and characterizing these genes in order to understand their molecular interactions and determine their role in pathogenesis. The black pigmentation of P. gingivalis is due to the accumulated hemin used as an iron source for growth. The organism appears to lack known siderophore activities and must use alternate mechanisms to sequester and transport exogenous iron. The expression of several outer membrane proteins is induced or repressed by heme, and a heme-repressible outer surface protein, which is translocated to the outer surface under heme-limiting conditions, is able to bind hemin (Bramanti and Holt, 1990; 1991; 1992; 1993). Genco et al., (1994) showed that hemin binding was induced by growth in hemin, a discrepancy which is attributed to differences in bacterial growth and assay conditions. P. gingivalis hemolytic activity is associated with the cell surface and outer membrane vesicles and two hemolysin genes have been cloned (Karunakaran and Holt, 1993). Hoover and Yoshimura (1994) reported the isolation of pigment-deficient (non-heme accumulating) transposon mutants also defective in trypsin-like protease activity. These data suggest that these virulence functions may be genetically linked. Using a similar screen, Genco et al. (1995b) observed that non-pigmented mutants had increased proteolytic activity. |
||||
|
This page is created and maintained by Drs. Margaret Duncan, Floyd Dewhirst, and Tsute Chen, Department of Molecular Genetics, The Forsyth Institute . Last modified on 02/20/2002 Copyright 2000, 2001, 2002 by The Forsyth Institute |